This interactive computational experiment aims at studying the influence of concentration and valency on the capacity of macromolecules to phase separate.
It was designed and implemented by Margot Riggi, under the supervision of Janet Iwasa and in collaboration with Ofer Rog (University of Utah). This work was funded through NSF grant 2219605.
We recommend visiting this webpage on a computer.

> A simulation of the system at the molecular scale, accompanied by a graph showing the evolution of the size of the clusters over time, normalized based on the total number of molecules present in the system.
> A schematic animation of the system at the macroscopic scale that reflects the experimental results from this study (Li & al, Nature, 2012).
Importantly, the animations may not always perfectly align with the results from the corresponding simulation at the molecular level. This could be due to various potential reasons, including unperfect parametrization of the simulations, or the fact that simulations are performed with a limited number of molecules.
Note that the time scale of both visualizations is also different.
The main points to remember from this experiment are summarized in the video at the bottom of the page.
1) Select the two conditions that you wish to compare by clicking on the corresponding dots in the plots below:
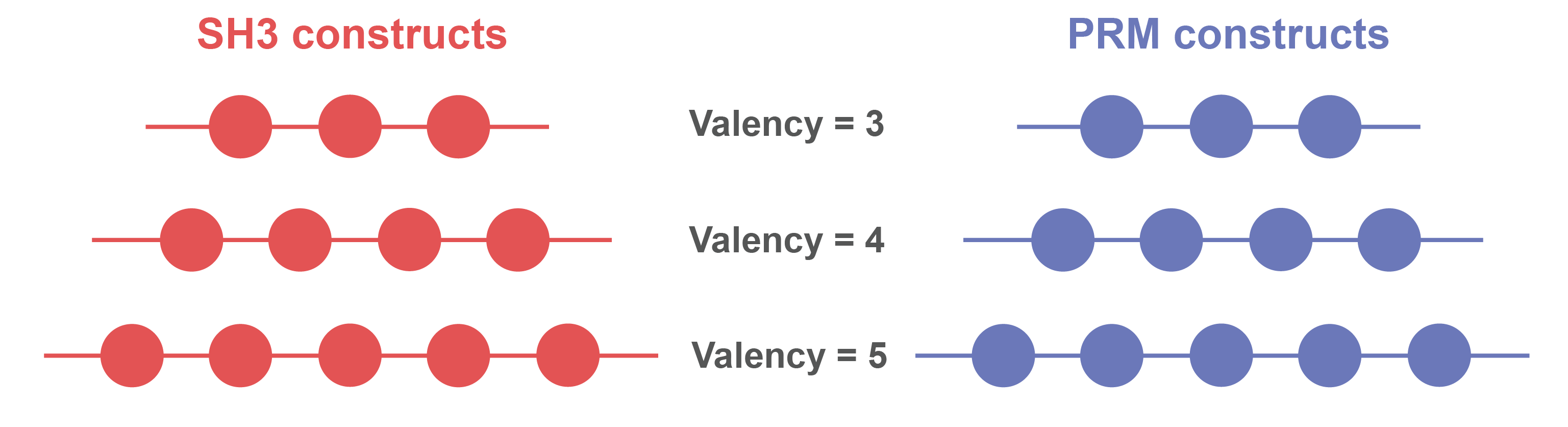
> Each plot corresponds to constructs of a different valency.
> Within a plot, each dot corresponds to a different combination of SH3 and PRM domain concentration.
2) Load the visualizations of the experiments by clicking on the corresponding button below the plots. Re-loading is required each time you change a condition.
3) The play button lets you start and pause both videos simultaneously.
Watch the video below for a summary of the main findings of this experiment!
Right click on the video to download.